Antibacterial Activity of Soil Bacteria against Escherichia coli and GC-MS Analysis of their Organic Compounds
Main Article Content
Abstract
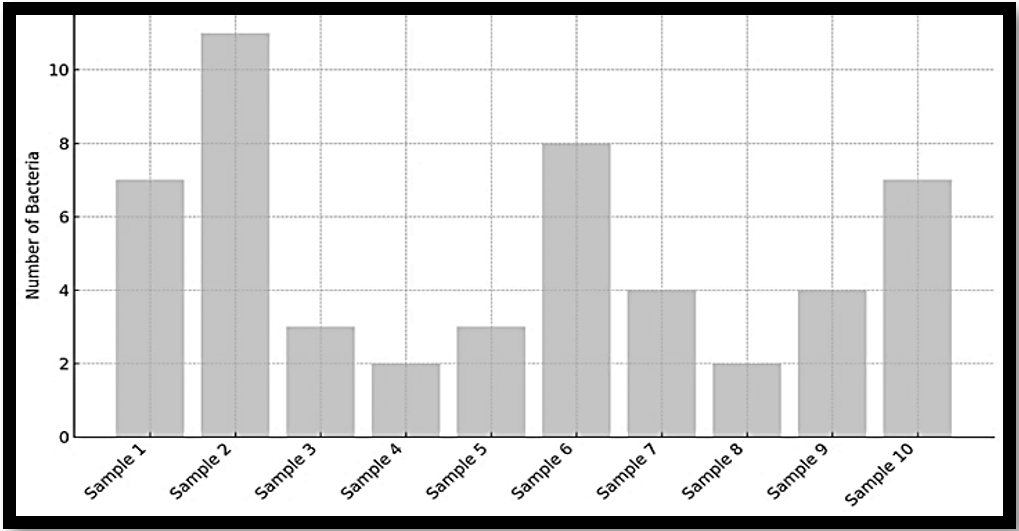
Antimicrobial agents encompass a wide range of compounds, including antibiotics, bacteriocins, and lipopeptides, which play a crucial role in combating infectious diseases. Antibiotics, in particular, are secondary metabolites of low molecular weight predominantly synthesized by soil dwelling microorganisms. These microbial metabolites have long served as a vital source of clinically important therapeutic agents. Members of the genus Bacillus and other rhizosphere-associated bacteria are especially known for producing diverse antimicrobial substances. In this study, ten rhizosphere soil samples were collected from different sites in Sana’a city, Yemen. From these samples, 50 antibiotic-producing soil bacteria were isolated. Bioactive metabolites were extracted using the solvent extraction method with chloroform and Ethanol. The crude extracts were analyzed by Gas Chromatography–Mass Spectrometry (GC–MS) to identify their organic composition. Among the 50 bacterial isolates, 14 showed antibacterial activity against resistant Escherichia coli using the agar well diffusion method. Further secondary screening revealed that the filtrates of four isolates Pseudomonas fluorescens, Bacillus subtilis, Acinetobacter baylyi, and Azotobacter vinelandii exhibited the strongest antibacterial effects. GC–MS analysis showed that each isolate produced more than eighty organic compounds; however, only a subset demonstrated antibacterial activity. The most notable bioactive compounds detected included Phenol, 4-(2-aminoethyl)- (CAS: tyramine), 2,4-di-tert-butylphenol, and n-hexadecanoic acid. This study highlights the potential of rhizosphere soil bacteria as promising sources of novel bioactive compounds. The identification of active metabolites and their antibacterial properties against resistant E. coli underscores their possible application in the development of alternative therapeutic strategiesto address antibiotic resistance.
Downloads
Article Details

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.
References
Aasfar A, Bargaz A, Yaakoubi K, Hilali A, Bennis I. Nitrogen‑fixing Azotobacter species as potential soil biological enhancers for crop nutrition and yield stability. Front Microbiol. 2021;12:628379. doi:10.3389/fmicb.2021.628379.
Ahmad F, Ahmad I, Khan M. Screening of free‑living rhizospheric bacteria for their multiple plant growth promoting activities. Microbiol Res. 2008;163(2):173–181. doi:10.1016/j.micres.2006.04.001.
Ahmed SS, Benhadj D, Lamari F. GC–MS analysis of bioactive metabolites from halotolerant bacteria isolated from the Sebkha of Oran (Algeria) and their antimicrobial properties. Microorganisms. 2021;9(10):2132. doi:10.3390/microorganisms9102132.
Ajilogba CF, Babalola OO. GC–MS analysis of volatile organic compounds from Bambara groundnut rhizobacteria and their antibacterial properties. World J Microbiol Biotechnol. 2019;35:83. doi:10.1007/s11274-019-2660-7
Al-Abdulsalam H, Almalki M, Khalifa A. Isolation and characterization of antimicrobial metabolites producing bacteria from soils in Al‑Ahsa, Saudi Arabia. J Microbiol Biotechnol Food Sci. 2025;e11779. doi:10.55251/jmbfs.11779.
Anantha Padmanabhan S, Deventhiran M, Saravanan P, Anand D, Rajarajan S. A comparative GC‑MS analysis of bacterial secondary metabolites of Pseudomonas species. Pharma Innov J. 2016;5(4):84–89.
Bahram M, Hildebrand F, Forslund SK, Anderson JL, Soudzilovskaia NA, Bodegom PM, et al. Structure and function of the global topsoil microbiome. Nature. 2018;560(7717):233–237. doi:10.1038/s41586-018-0386-6.
Berdy J. Bioactive microbial metabolites. J Antibiot (Tokyo). 2005;58(1):1–26. doi:10.1038/ja.2005.1.
Cappuccino JG, Sherman N. Microbiology: A Laboratory Manual. 10th ed. Pearson Education; 2014.
Cappuccino JG, Welsh C. Microbiology: A Laboratory Manual. 11th ed. Pearson; 2017.
Caulier S, Nannan C, Gillis A, Licciardi F, Bragard C, Mahillon J. Overview of the antimicrobial compounds produced by members of the Bacillus subtilis group. Front Microbiol. 2019;10:302. doi:10.3389/fmicb.2019.00302
Centers for Disease Control and Prevention (CDC). Antibiotic Resistance Threats in the United States [Internet]. 2023 [cited 2025 Jun 7]. Available from: https://www.cdc.gov/antimicrobial-resistance/
Chen W, Gong L, Guo Z, Wang W. Antibacterial properties of plant essential oils and their components against foodborne pathogens. J Agric Food Chem. 2017;65(4):720–728. doi:10.1021/acs.jafc.6b04250
Desbois AP, Smith VJ. Antibacterial free fatty acids: activities, mechanisms of action and biotechnological potential. Prog Lipid Res. 2010;49(1):1–13. doi:10.1016/j.plipres.2009.08.003
Friedman M. Antibacterial, antiviral, and antifungal properties of caffeine and related methylxanthines. Food Chem. 2017;221:453–464. doi:10.1016/j.foodchem.2016.10.109.
González H, et al. Green extraction of secondary metabolites from plants: obstacles, current status, and trends. Sustain Chem Environ. 2024;8:100157. doi:10.1016/j.scenv.2024.100157.
Javed H, Riaz A, Qureshi A, Javed K, Mujeeb F, Ijaz F, et al. Isolation,characterization and screening of PGPR capable of providing relief in salinity stress. Eurasian J Soil Sci. 2020;9(2):85–91. doi:10.18393/ejss.650546.
Kaaria P, Matiru V, Ndungu M. Antimicrobial activities of secondary metabolites produced by endophytic bacteria from selected indigenous Kenyan plants. Afr J Microbiol Res. 2012;6(45):7253–7258.
Kai M. Diversity and distribution of volatile secondary metabolites throughout Bacillus subtilis isolates. Front Microbiol. 2020;11:559. doi:10.3389/fmicb.2020.00559.
Khan MA, Yasien S, Iqbal MM, Javed M. Comparative evaluation of various extraction techniques for secondary metabolites from Bombax ceiba L. flowering plants along with in vitro anti‑diabetic performance. Bioengineering. 2022;9(10):486. doi:10.3390/bioengineering9100486.
Numan M, Shah M, Asaf S, Rehman NU, Al-Harrasi A. Bioactive compounds from endophytic bacteria Bacillus subtilis strain EP1 with their antibacterial activities. Metabolites. 2022;12(12):1228. doi:10.3390/metabo12121228.
Kovács A, Nagy R. Assessment of antibacterial potential using the agar well diffusion method. Eur J Microbiol Immunol. 2024;14(2):123–130. doi:10.1556/1886.2024.00015.
Król E, Gąsiorowski K, Mucha P. Antibacterial and antifungal activity of cyclic siloxanes. Microbiol Biotechnol. 2017;101(11):4419–4433. doi:10.1007/s00253-017-8232-5.
Kumar S, Sharma A, Verma A. Identification of bioactive secondary metabolites from Bacillus species by GC–MS profiling. J Appl Microbiol. 2022;134(2):453–468. doi:10.1111/jam.14999.
Kurrey NK, Singh S, Yadav AN. Azotobacter: a potential bio‑fertilizer for soil and plant health management. Saudi J Biol Sci. 2020;27(12):3634–3640. doi:10.1016/j.sjbs.2020.09.054.
Li Y, Xiong W, Wang Y, Sun Y, Zeng Q. Metabolic capabilities of Acinetobacter baylyi in the degradation of pollutants: implications for bioremediation. Front Microbiol. 2020;11:615. doi:10.3389/fmicb.2020.00615.
Mahfouz N, Ferreira I, Beisken S, von Haeseler A, Heringa J. Antimicrobial resistance surveillance using machine learning methods. Nat Commun. 2024;15:1512. doi:10.1038/s41467-024-42021-7.
Manga BS, Oyeleke SB. Essentials of M. Higton, Industrial Microbiology: a laboratory practical’s in microbiology. 1st ed. London: Blackwell Publisher/Tobes; 2008. p.56–76.
Musliu A, Salawudeen W. Screening and isolation of the soil bacteria for ability to produce antibiotics. Eur J Appl Sci. 2012;4(5):211–215. doi:10.5829/idosi.ejas.2012.4.5.2011.
Nakatsuji T. Antimicrobial property of lauric acid against Propionibacterium acnes: its therapeutic potential for inflammatory acne vulgaris. J Investig Dermatol. 2009;129(10):2480–2488. doi:10.1038/jid.2009.93.
Qin S, Xiao W, Zhou C, Pu Q, Deng X, Lan L, et al. Pseudomonas aeruginosa: pathogenesis, virulence factors, antibiotic resistance, interaction with host, technology advances and emerging therapeutics. Signal Transduct Target Ther. 2022;7(1):199. doi:10.1038/s41392-022-01084-9.
Qu Q, Zhang ZY, Peijnenburg WJGM, Liu WY, Lu T, Hu BL, et al. Rhizosphere microbiome assembly and its impact on plant growth. J Agric Food Chem. 2020;68(18):5024–5038. doi:10.1021/acs.jafc.0c00073.
Santala S, Santala V. Acinetobacter baylyi ADP1—naturally competent for synthetic biology. Essays Biochem. 2021;65(2):309–318. doi:10.1042/EBC20200168.
Shaikh S, Fatima J, Shakil S, Rizvi SMD, Kamal MA. Antibiotic resistance and extended-spectrum beta-lactamases: Types, epidemiology and treatment. Saudi J Biol Sci. 2023;30(1):103448. doi:10.1016/j.sjbs.2022.103448.
Sorg O, Antille C, Kaya G, Saurat JH. Retinoids in cosmeceuticals. J Steroid Biochem Mol Biol. 2006;101(4–5):136–141. doi:10.1016/j.jsbmb.2006.09.033.
Sumbul A, Tiwari A, Khare E. Volatile organic compounds from Pseudomonas fluorescens: Mechanisms of plant growth promotion and pathogen suppression. World J Microbiol Biotechnol. 2023;39:49. doi:10.1007/s11274-023-03873-0.
Tiwari K, Gupta RK. Rare actinomycetes: A potential storehouse for novel antibiotics. Crit Rev Biotechnol. 2012;32(2):108–132. doi:10.3109/07388551.2011.562482.
Todar K, Ubukata M, Hamada M. Microbiology: A human perspective. London: McGraw-Hill; 2005.
Ventola CL. The antibiotic resistance crisis: part 1: causes and threats. Pharm Ther. 2023;48(2):71–77. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC9876086/
World Health Organization (WHO). Global Antimicrobial Resistance and Use Surveillance System (GLASS) Report 2024. Available from: https://www.who.int/publications/i/item/9789240070650
Xun W, Yan R, Ren Y, Jin D, Xiong W, Zhang G, et al. Grazing-induced microbiome alterations drive soil organic carbon turnover and productivity in meadow steppe. Microbiome. 2018;6:170. doi:10.1186/s40168-018-0554-6
Zhang Y, Wang Q, Yao H, Wang Y. Pyoverdine production in Pseudomonas fluorescens is influenced by carbon source availability. J Basic Microbiol. 2020;60(6):570–8. Available from: https://pubmed.ncbi.nlm.nih.gov/32500426
Zhao J, Wang Y, Li L. GC–MS-based volatile organic compounds profile of Bacillus species and their pharmacological significance. Molecules. 2020;25(22):7556. doi:10.3390/molecules25227556